Projects overview
Prostate Cancer
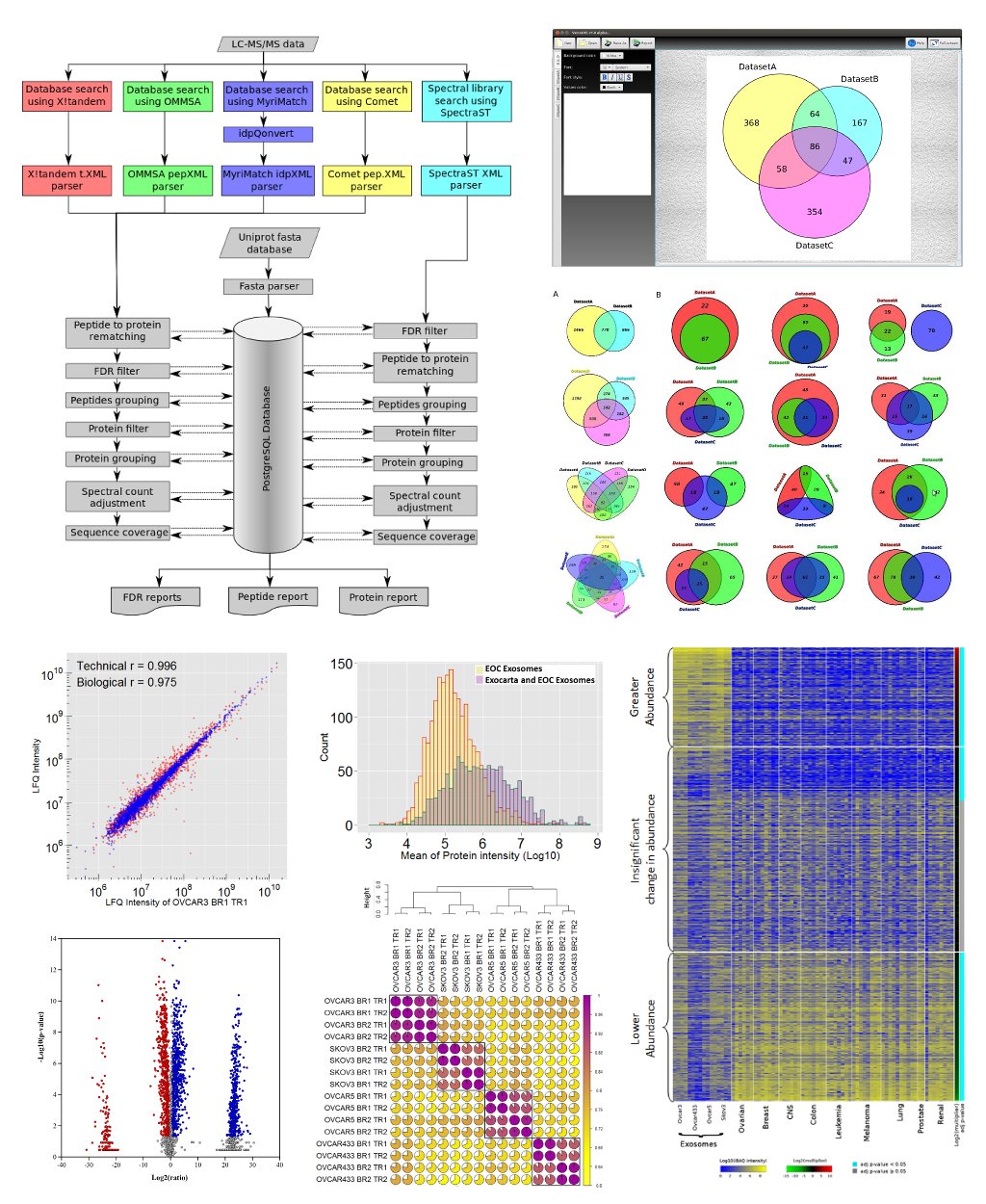
The main focus of our prostate cancer work is to use prostate proximal fluids (expressed prostatic secretions and post-DRE urines) to discover novel prostate cancer biomarkers. Initially, comprehensive proteome profiles were generated from richly annotated prostate proximal fluids. Computational data mining identified candidate peptides for the systematic development of targeted proteomics assays (Selected Reaction Monitoring Mass Spectrometry; SRM-MS). The goal of this work is to develop prognostic signatures to distinguish patients with aggressive and indolent prostate cancers. We have currently developed >100 SRM-MS assays and evaluating their prognostic relevance in clinically stratified post-DRE urines.
Team members: Yunee Kim (PhD student)
Head and Neck Cancer
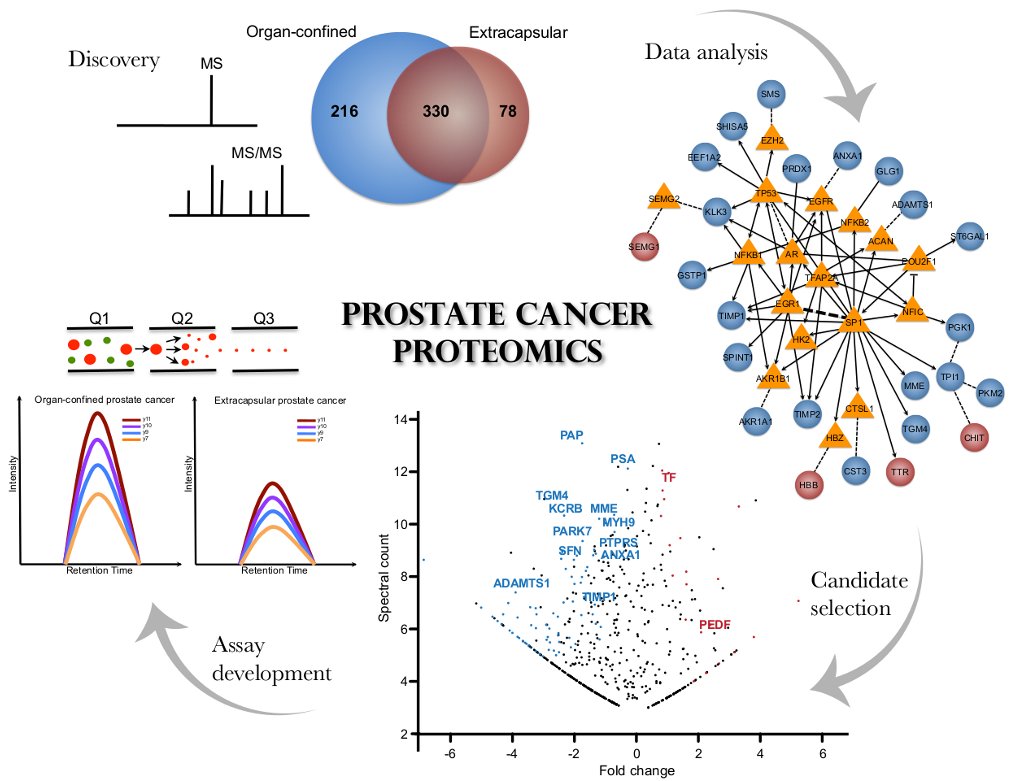
We currently have a variety of head and neck cancer proteomics projects in the lab. These include the detailed analyses of archival FFPE tissues from oropharyngeal tumors to identify molecular pathways based on HPV-status and to identify genomic and proteomics signatures that predict the development of metastasis.
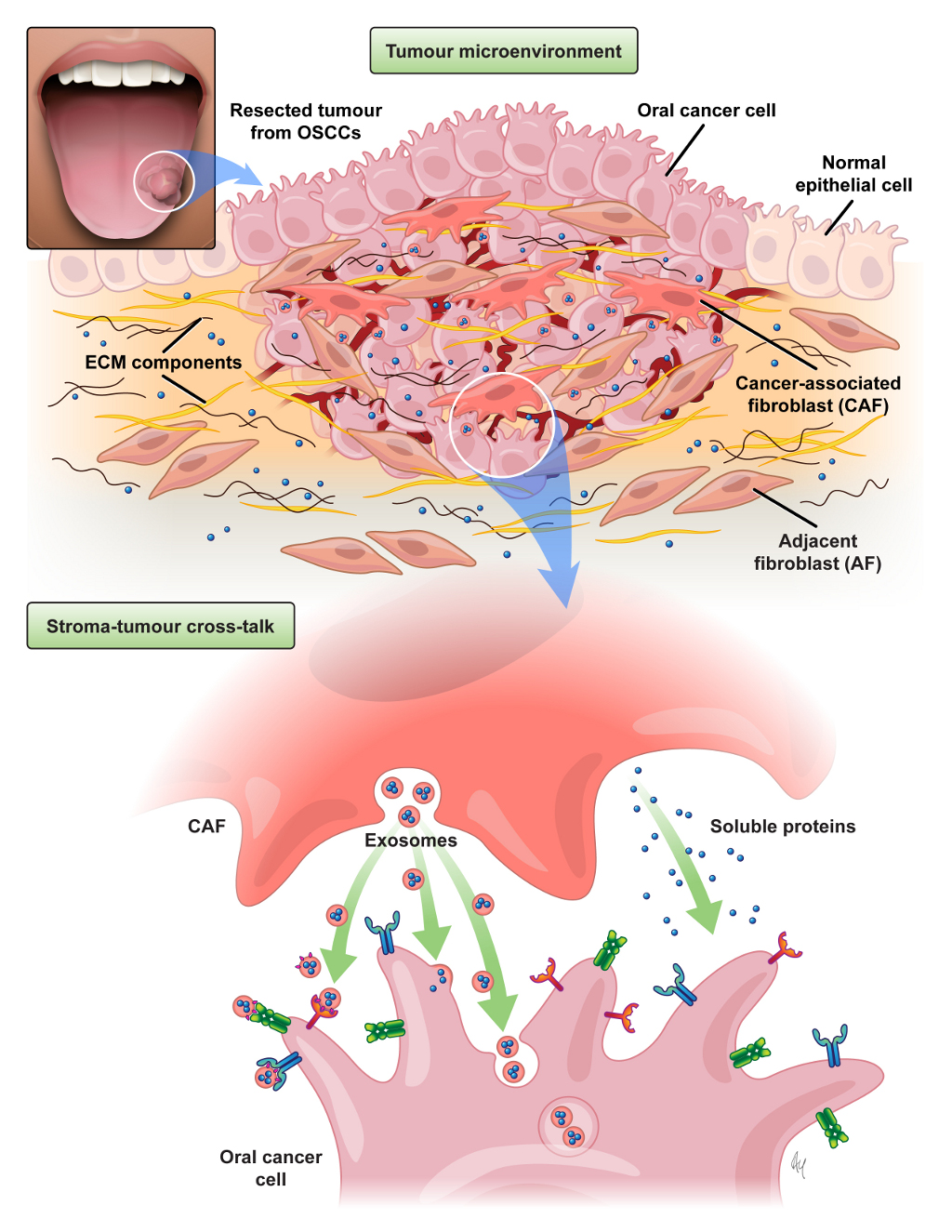
Additional projects include the identification of signaling pathways within the tumor microenvironment that support cancer progression, as well as the development and application of onco-proteogenomics analyses platforms. Briefly, a proteomic approach is used to identify proteins secreted from cancer-associated fibroblast (CAFs) within the tumor-stroma niche that can act as paracrine regulators of oral cancer growth. In this experimental model, the in-depth proteomic analysis of fibroblast secretomes and the cargo of extracellular nanovesicles are integrated with plasma membrane proteomes of oral cancer cells. Our goals are to provide insights into the functional role of a pro-invasive stroma and a mechanistic link between complex microenvironmental stimuli and phenotypic tumor response.
The complexity of the tumour genome and transcriptome are rapidly being uncovered by next-generation sequencing studies. But the way they are regulated and expressed into a functional proteome remains poorly understood. Standard proteomic workflows use databases generated from known proteins to identify peptides within complex mixtures. But these databases do not capture the uniqueness of the cancer transcriptome, with its point-mutations, unusual splice variants and gene fusions. There is a need to develop proteomic approaches that exploit individual-tumor genomic profiles. Onco-proteogenomics is aimed to integrate mass-spectrometry-generated proteomics data with genomic information to better characterize tumor phenotypes.
Team members: Simona Principe (post-doc), Lusia Sepiashvili (PhD student), Javier Alfaro (PhD student), Salvador Mejia (Research Associate)
Ovarian Cancer
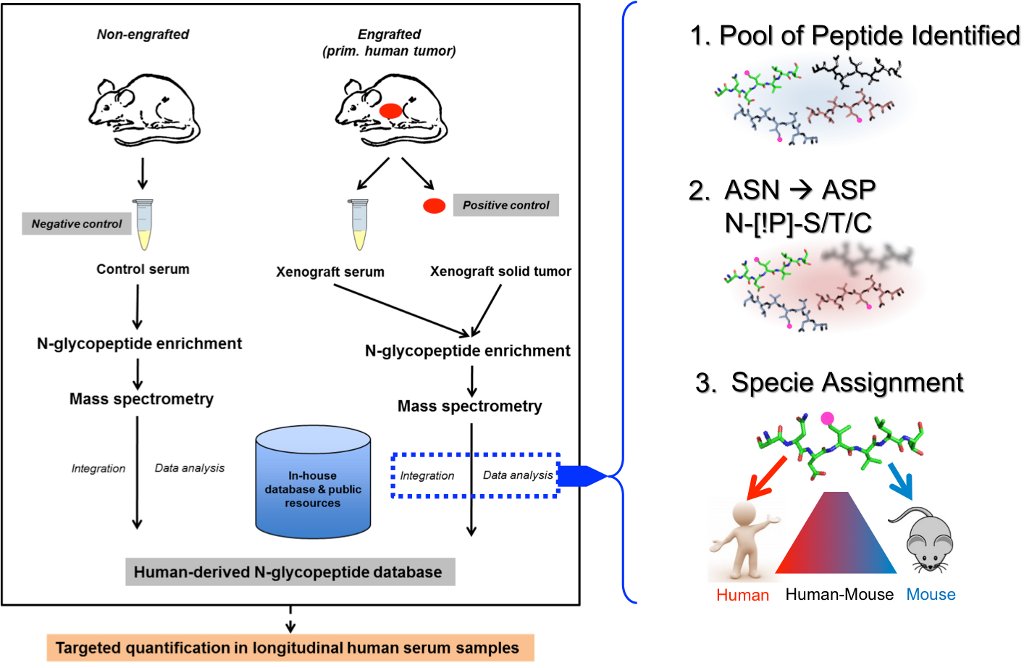
Our ovarian cancer project applies a N-glycoproteomics approach to systematically analyze sera and tissues of high-grade serous ovarian cancer xenograft models established from primary patient-derived tumors. We have developed a magnetic-bead-based approach using hydrazide chemistry to isolate N-glycosylated peptides. The goal is to identify proteins specifically secreted by tumor tissue and detected in serum samples that could be used as biomarkers for the detection of ovarian cancer recurrence (similar to CA125). Systematic bioinformatics ranking schemes, based on sequence and abundance will be applied to select the most reliable proteins markers for validation in human serum samples. Additional goals include correlating proteomic profile for subtypes of HGSOC tissues with available mRNA expression analysis. For these analyses a protocol has been developed that enriches for N-glycoproteins, phosphoproteins and total protein from primary tumor xenografts.
Team members: Ankit Sinha (PhD student)
Plasma membrane and exosome proteomics
Mammalian cells are composed of distinct cellular fractions, called organelles, performing precise cellular functions. Our lab has a long-standing interest in combining cellular fractionation with in-depth proteomics analyses. We are particularly interesting in developing and applying methodologies for the analyses of membrane proteins and extracellular membrane vesicles.
In the context of plasma membrane proteomics we are applying various direct labeling approaches, including silica-bead coating, cell-surface biotinylation and cell-surface capture (i.e. glyco-capture) to investigate dynamic changes in the plasma membrane proteome of cancer cell lines and cardiac-related cells and tissues.
The broad term "extracellular vesicles" summarizes a group of nanometer sized, membranous vesicles secreted from a variety of cell types. The most commonly studied vesicles include exosomes, microvesicles, prostasomes and apoptotic bodies. These vesicles differ in size, biogenesis, marker composition and molecular cargo such as DNA, RNA, proteins and lipids. Although the first manuscripts on exosomes date back nearly 30 years, the field of extracellular vesicle research has witnessed a significant surge in interest in recent years. We are interested in developing better proteomics methods for the analyses of extracellular vesicles and for the investigation of their biological roles in cancer biology.
Team members: Simona Principe (post-doc); Lusia Sepiashvili (PhD student), Ankit Sinha (PhD student) Salvador Mejia (technician)
Bioinformatics
Our bioinformatics team is involved in all projects listed above, providing data management and statistical analyses for proteomics data. We have implemented an in-house mass-spectrometry pipeline that incorporates multiple search algorithms (X!Tandem, OMSSA, MyriMatch, Comet) into a Python and PostgreSQL pipeline. In addition, a MaxQuant server is available for label-free quantitative proteomics. All our statistical analyses and visualizations are performed the open-source R pipeline. The team also uses various programming languages such us PHP/Perl/Java/C for biological data mapping and presentation.
Software: VennDIS: A JavaFX-based Venn and Euler Diagram Software to Generate Publication Quality Figures.
Team members: Alex Ignatchenko (programmer), Vladimir Ignatchenko (programmer), Javier Alfaro (PhD student), Ankit Sinha (PhD student)